Photo by Pawel Czerwinski on Unsplash
Sonic Log Prediction
Summary
Sonic logs are required for well-seismic ties, an essential step in the exploration workflow. However, due to various resource constraints, sonic logs might not be recorded in several wells in a field. On the other hand, it is seen that basic logs like Gamma Ray, Neutron Porosity, Bulk Density, Resistivity are almost always recorded in all wells. Geoscientists approximate missing sonic logs from sonic logs of nearby wells using empirical techniques. These techniques might not generalise to wells other than the well in question. There is also the consistency problem.
This analysis uses a data driven approach to predict missing sonic logs on a field scale. This might help predict sonic logs for all wells in a field , in turn vastly improving well seismic ties, seismic resolution , depth migration etc. This Analysis predicts Delta-T compressional from GR, RHOB, NPHI using Machine learning based algorithms. The dataset consists of 8 wells from Poseidon field of North West Australian Offshore, obtained from Data Underground. LAS files containing spliced well logs sampled at 0.5m interval are used for the analysis ( Spliced logs provided by Occam Technologies ).
3 Models have been trained, one each using Support Vector Machines, Gradient Boosting & Artificial Neural Networks.
The model is trained on 6 out of 8 wells and validated on 2 wells.
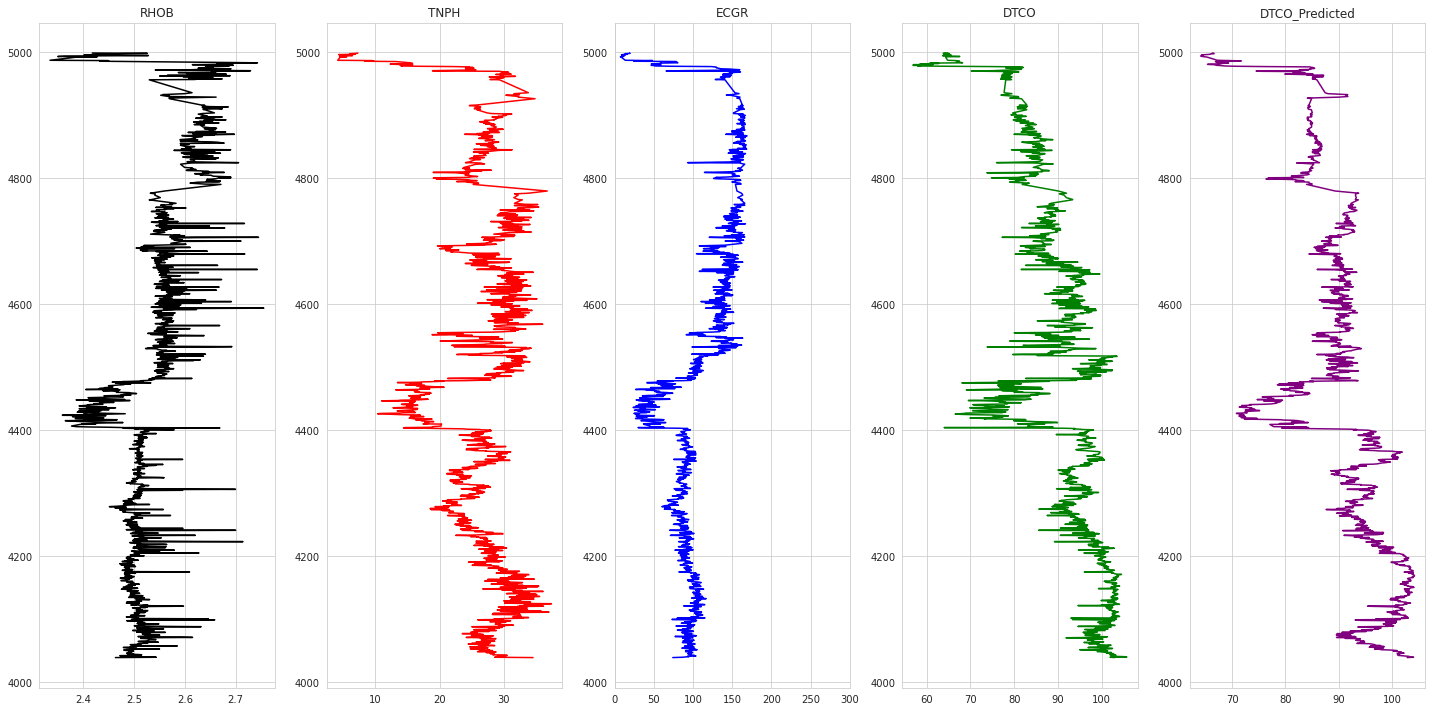
The analysis shows promising results with the mean square error obtained on validation wells being 0.165 & 0.07. Further, predicted sonic logs have high negative correlation with bulk density as dictated by physics.
Importing Data
1import numpy as np2import pandas as pd3import glob4import lasio as ls5import matplotlib.pyplot as plt6import warnings7warnings.filterwarnings('ignore')
1log_list = glob.glob('Poseidon_logs/*.LAS')2log_list
1['Poseidon_logs/Poseidon1Decim.LAS',2 'Poseidon_logs/Kronos1Decim.LAS',3 'Poseidon_logs/Boreas1Decim.LAS',4 'Poseidon_logs/Proteus1Decim.LAS',5 'Poseidon_logs/Pharos1Decim.LAS',6 'Poseidon_logs/Torosa1LDecim.LAS',7 'Poseidon_logs/PoseidonNorth1Decim.LAS',8 'Poseidon_logs/Poseidon2Decim.LAS']
1# Predictors2columns_of_interest = ['RHOB', 'TNPH','ECGR','DTCO']3mnemonic_replacements = {4 'NPHI' : 'TNPH' ,5 'HROM' : 'RHOB',6 'GR' : 'ECGR',7 'GRD' : 'ECGR',8 'TNP' : 'TNPH',9 'RHOZ' : 'RHOB',10 'HTNP' : 'TNPH',11 'BATC' : 'DTCO',12 'GRARC' : 'ECGR',13 'DTC' : 'DTCO'1415}
1# Reading LAS files23las_data = {}4las_df = {}5well_list = []67for las_file in log_list :8 well_name = las_file.split('Decim.LAS')[0].split('/')[1]910 well_list.append(well_name)1112 las = ls.read(las_file)1314 # saving las in las_data1516 las_data[well_name] = las1718 df = las.df()1920 # mnemonic replacements21 for column in df.columns.values :22 if column in mnemonic_replacements.keys() :23 df = df.rename(columns = {column : mnemonic_replacements[column]})242526 las_df[well_name] = df27 print('\nWell Name : ', well_name )28 print(df.head())29 print("No of datapoints :",df.shape[0])30print(well_list)
1Well Name : Poseidon12 ECGR ATRX ATRT TNPH CAL1 HDAR RHOB DTSM DTCO3DEPT4579.5 NaN 0.6000 0.6700 NaN NaN NaN NaN NaN NaN5580.0 NaN 0.6700 0.7000 NaN NaN NaN NaN NaN NaN6580.5 NaN 0.6200 0.6700 NaN NaN NaN NaN NaN NaN7581.0 NaN 0.5500 0.6400 NaN NaN NaN NaN NaN NaN8581.5 NaN 0.5598 0.6295 NaN NaN NaN NaN NaN NaN9No of datapoints : 90731011Well Name : Kronos112 ECGR RS RD TNPH RHOB DCAV DTCO DTSM13DEPT14531.0 0.5559 NaN NaN NaN NaN NaN NaN NaN15531.5 0.9649 NaN NaN NaN NaN NaN NaN NaN16532.0 0.7309 NaN NaN NaN NaN NaN NaN NaN17532.5 4.3208 NaN NaN NaN NaN NaN NaN NaN18533.0 -0.0001 NaN NaN NaN NaN NaN NaN NaN19No of datapoints : 95842021Well Name : Boreas122 ECGR RS RD RHOB TNPH DTCO DTSM HDAR23DEPT24200.0 3.8287 NaN NaN NaN NaN NaN NaN NaN25200.5 3.8287 NaN NaN NaN NaN NaN NaN NaN26201.0 3.1437 NaN NaN NaN NaN NaN NaN NaN27201.5 1.4981 NaN NaN NaN NaN NaN NaN NaN28202.0 3.9696 NaN NaN NaN NaN NaN NaN NaN29No of datapoints : 100123031Well Name : Proteus132 ECGR RS RD DTCO DTSM HDAR RHOB TNPH33DEPT34489.5 16.2084 NaN NaN NaN NaN NaN NaN NaN35490.0 20.6540 NaN NaN NaN NaN NaN NaN NaN36490.5 19.2559 NaN NaN NaN NaN NaN NaN NaN37491.0 15.8395 NaN NaN NaN NaN NaN NaN NaN38491.5 14.7239 NaN NaN NaN NaN NaN NaN NaN39No of datapoints : 95214041Well Name : Pharos142 RHOB TNPH ECGR P16H P34H DTCO DTS43DEPT44482.0 NaN NaN 48.9556 NaN NaN NaN NaN45482.5 NaN NaN 54.2686 NaN NaN NaN NaN46483.0 NaN NaN 56.0523 NaN NaN NaN NaN47483.5 NaN NaN 53.8891 NaN NaN NaN NaN48484.0 NaN NaN 67.2855 NaN NaN NaN NaN49No of datapoints : 94885051Well Name : Torosa1L52 ECGR RS RD RHOB TNPH HDAR DTCO DTS53DEPT54555.0 12.3003 0.6826 0.7422 NaN NaN NaN NaN NaN55555.5 24.5442 0.4900 0.7223 NaN NaN NaN NaN NaN56556.0 25.4925 0.4401 0.4498 NaN NaN NaN NaN NaN57556.5 25.2527 0.4874 0.7347 NaN NaN NaN NaN NaN58557.0 28.6476 0.4607 0.6694 NaN NaN NaN NaN NaN59No of datapoints : 82576061Well Name : PoseidonNorth162 ECGR RS RD RHOB TNPH DTCO DTSM HDAR63DEPT64508.0 24.4403 NaN NaN NaN NaN NaN NaN NaN65508.5 29.6471 NaN NaN NaN NaN NaN NaN NaN66509.0 36.3076 NaN NaN NaN NaN NaN NaN NaN67509.5 56.3806 NaN NaN NaN NaN NaN NaN NaN68510.0 54.7321 NaN NaN NaN NaN NaN NaN NaN69No of datapoints : 95447071Well Name : Poseidon272 ECGR RS RD DTCO DTSM DCAV RHOB TNPH73DEPT74490.0 17.6006 0.6898 0.3094 NaN NaN NaN NaN NaN75490.5 17.6006 0.6898 0.3094 NaN NaN NaN NaN NaN76491.0 17.6006 0.6898 0.3094 NaN NaN NaN NaN NaN77491.5 16.7792 0.6959 0.3141 NaN NaN NaN NaN NaN78492.0 15.9579 0.6959 0.3141 NaN NaN NaN NaN NaN79No of datapoints : 972380['Poseidon1', 'Kronos1', 'Boreas1', 'Proteus1', 'Pharos1', 'Torosa1L', 'PoseidonNorth1', 'Poseidon2']
The well names are : ‘Boreas1’, ‘Poseidon1’, ‘Kronos1’, ‘Proteus1’, ‘Poseidon2’, ‘Torosa1L’, ‘PoseidonNorth1’, ‘Pharos1’.
The following curves are extracted from each LAS file.
- ECGR - EDTC Corrected Gamma Ray
- RHOB - Bulk Density
- TNPH - Thermal Neutron Porosity ( Ratio Method ) in selected lithology
- DTCO - Delta T Compressional
Visualising Logs
1# Plotting Logs2def plot_logs(well) :34 fig,ax = plt.subplots(nrows=1, ncols= len(columns_of_interest), figsize=(20,10))5 colors = ['black', 'red', 'blue', 'green', 'purple', 'black', 'orange']67 for i,log in enumerate(columns_of_interest) :8 ax[i].plot(well[log], well['DEPT'], color = colors[i])9 ax[i].set_title(log)10 ax[i].grid(True)1112 ax[2].set_xlim(0,300)13 plt.tight_layout(1.1)14 plt.show()1516for well in well_list :17 print('Well Name :', well)18 well1 = las_df[well]19 plot_logs(well1[columns_of_interest].reset_index())
1Well Name : Poseidon1

1Well Name : Kronos1
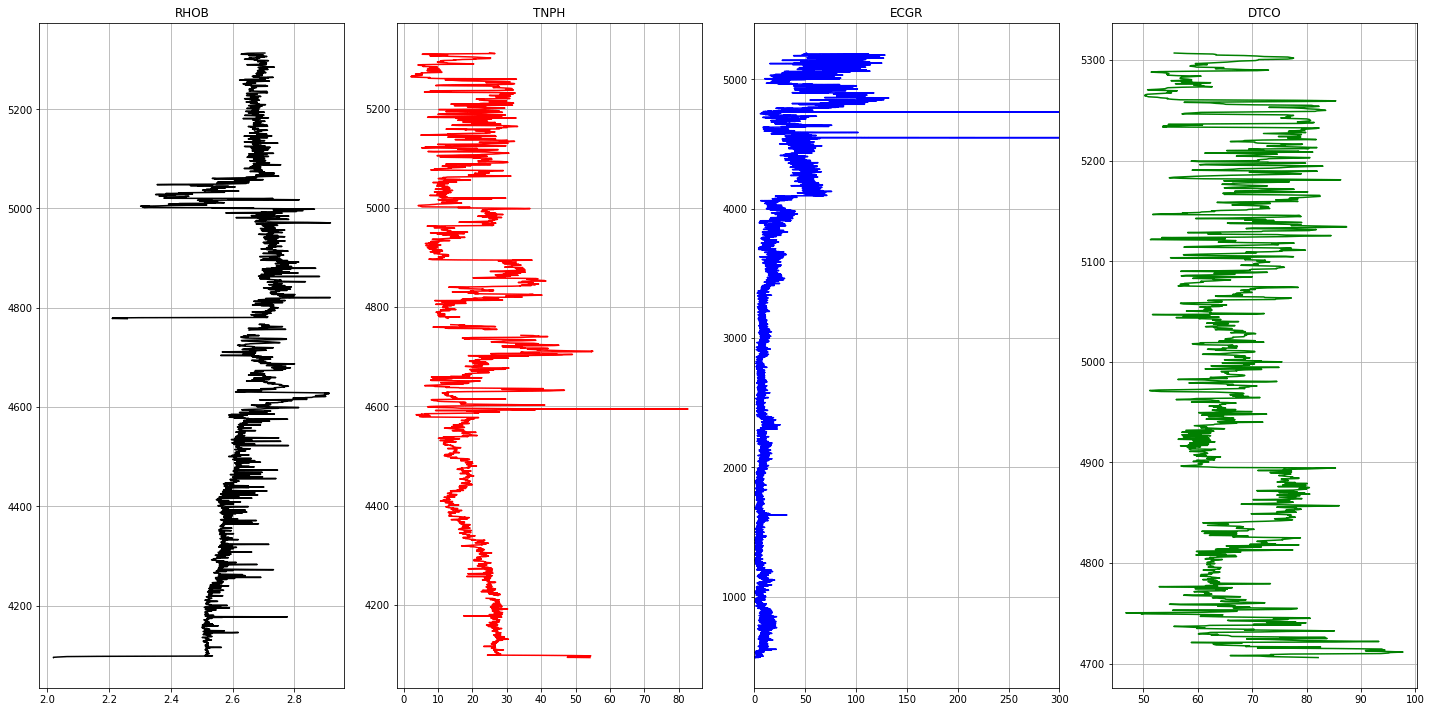
1Well Name : Boreas1
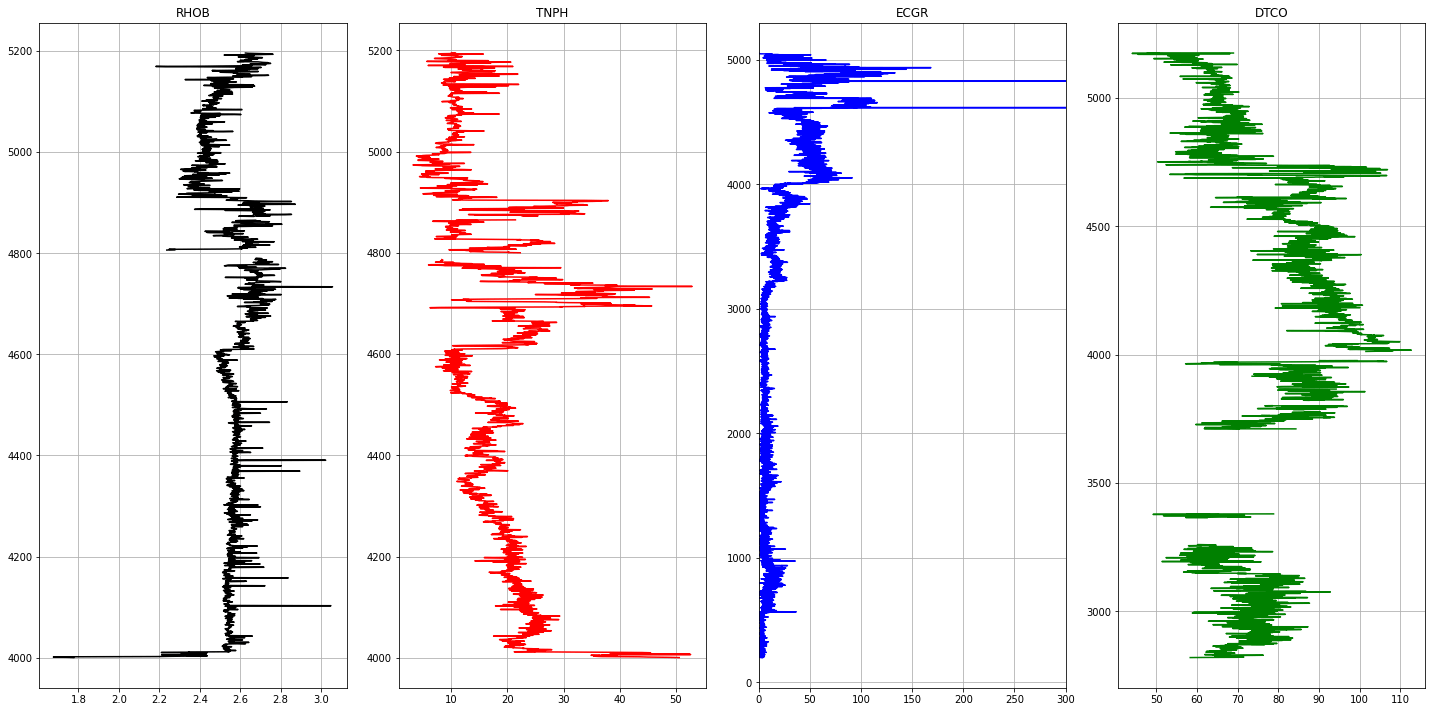
1Well Name : Proteus1
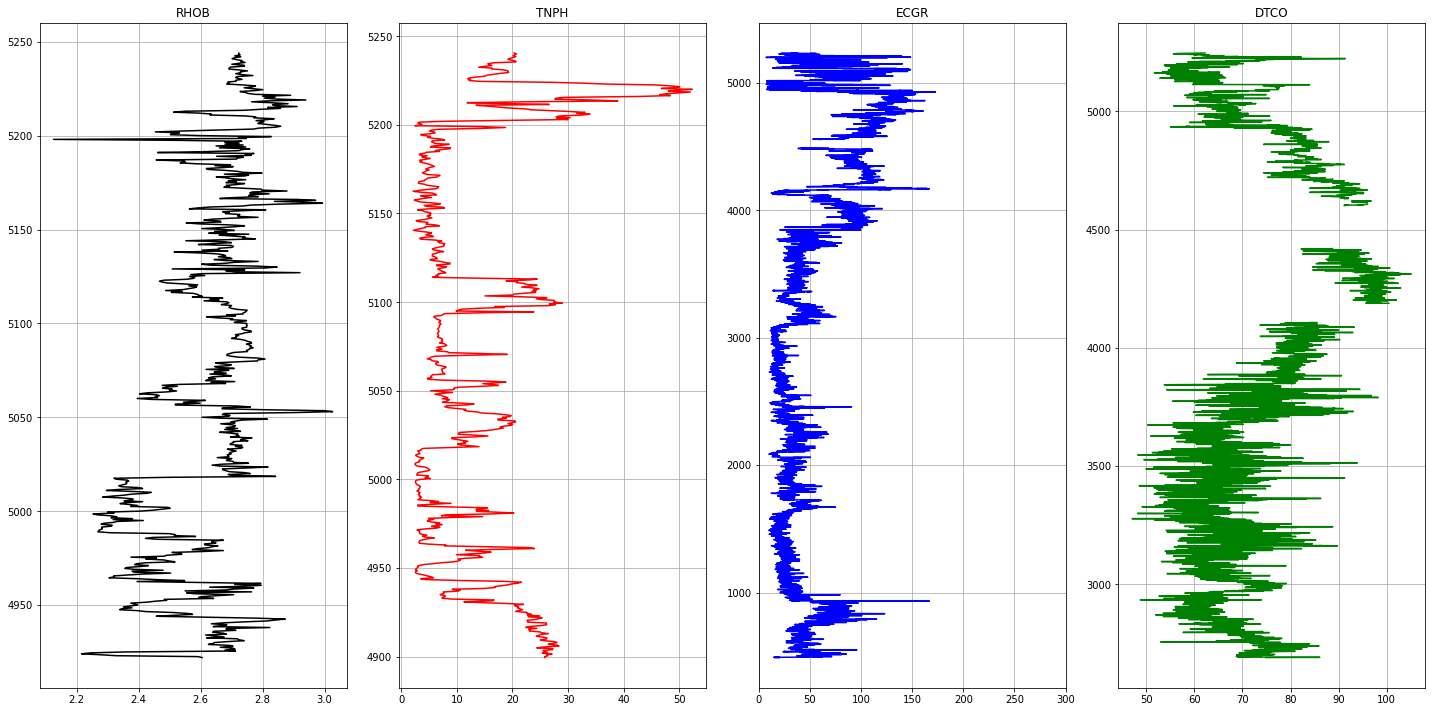
1Well Name : Pharos1
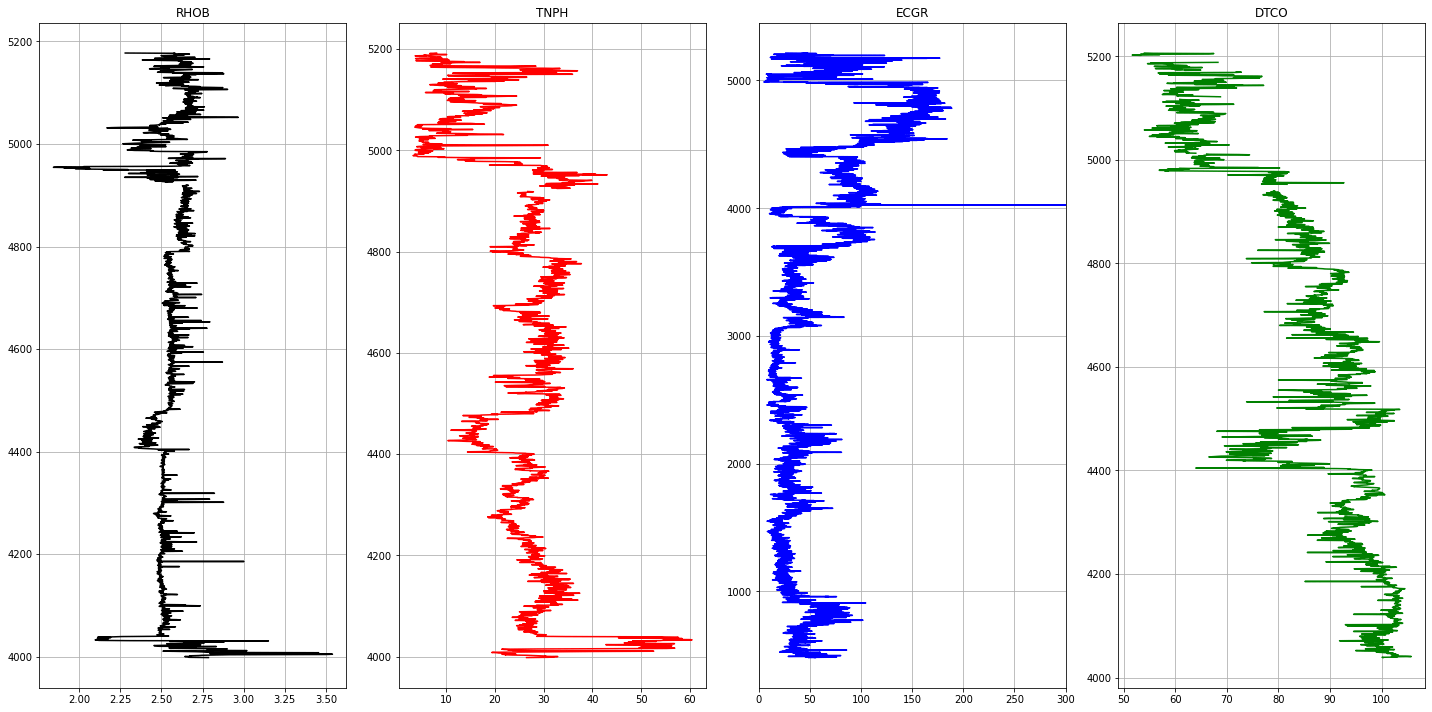
1Well Name : Torosa1L
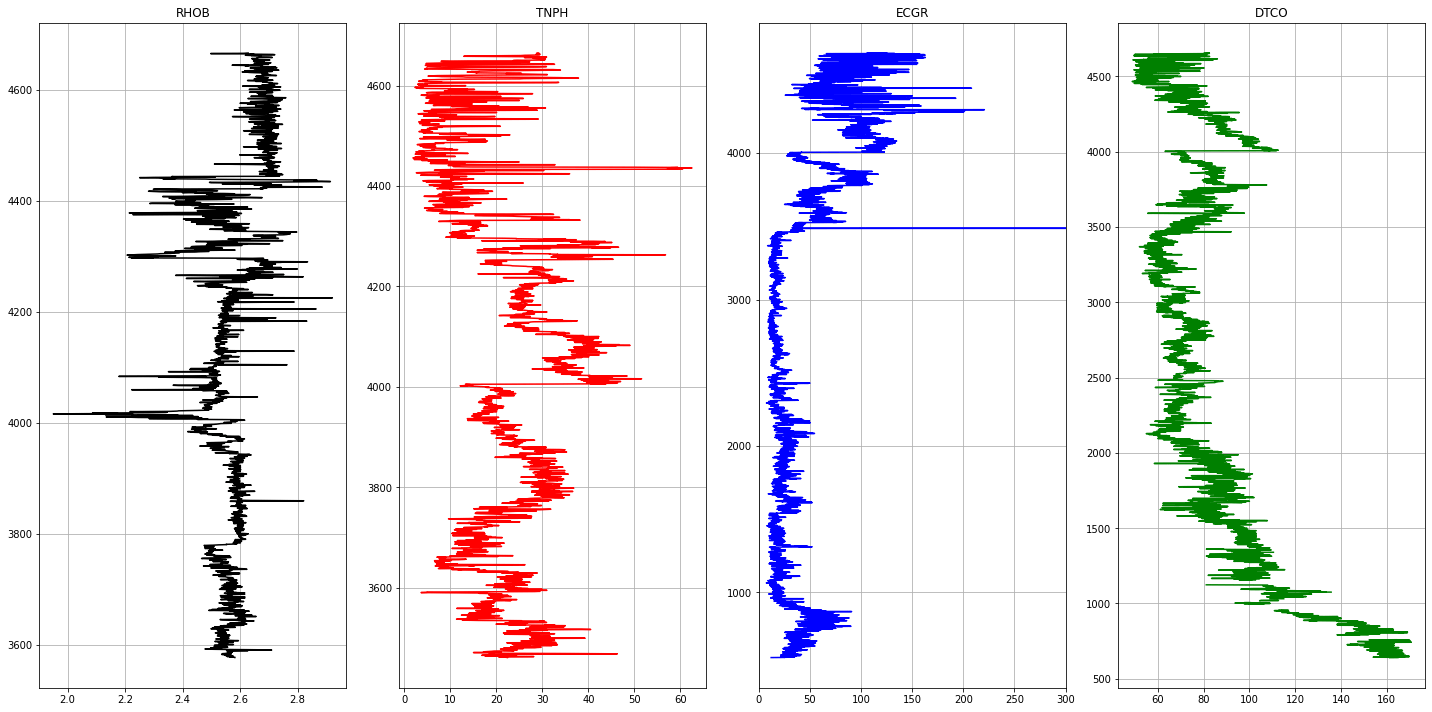
1Well Name : PoseidonNorth1
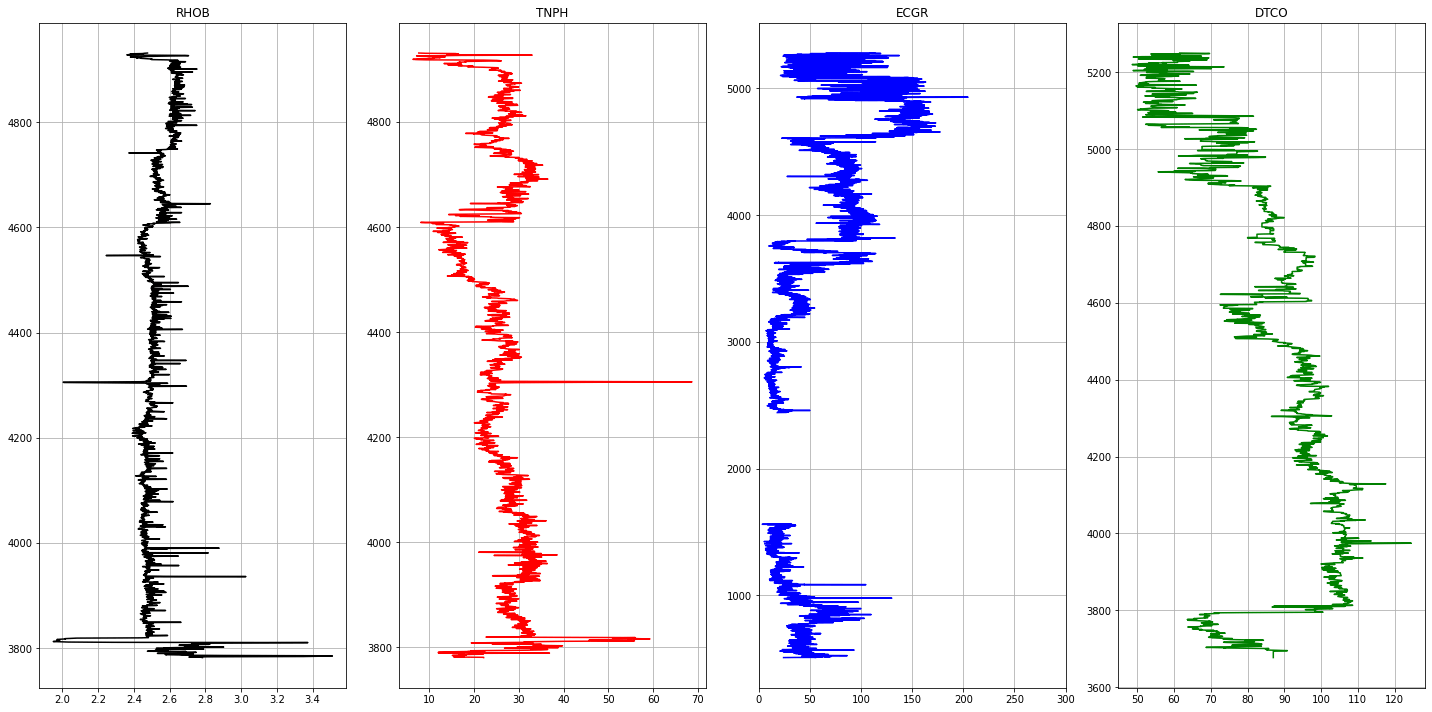
1Well Name : Poseidon2
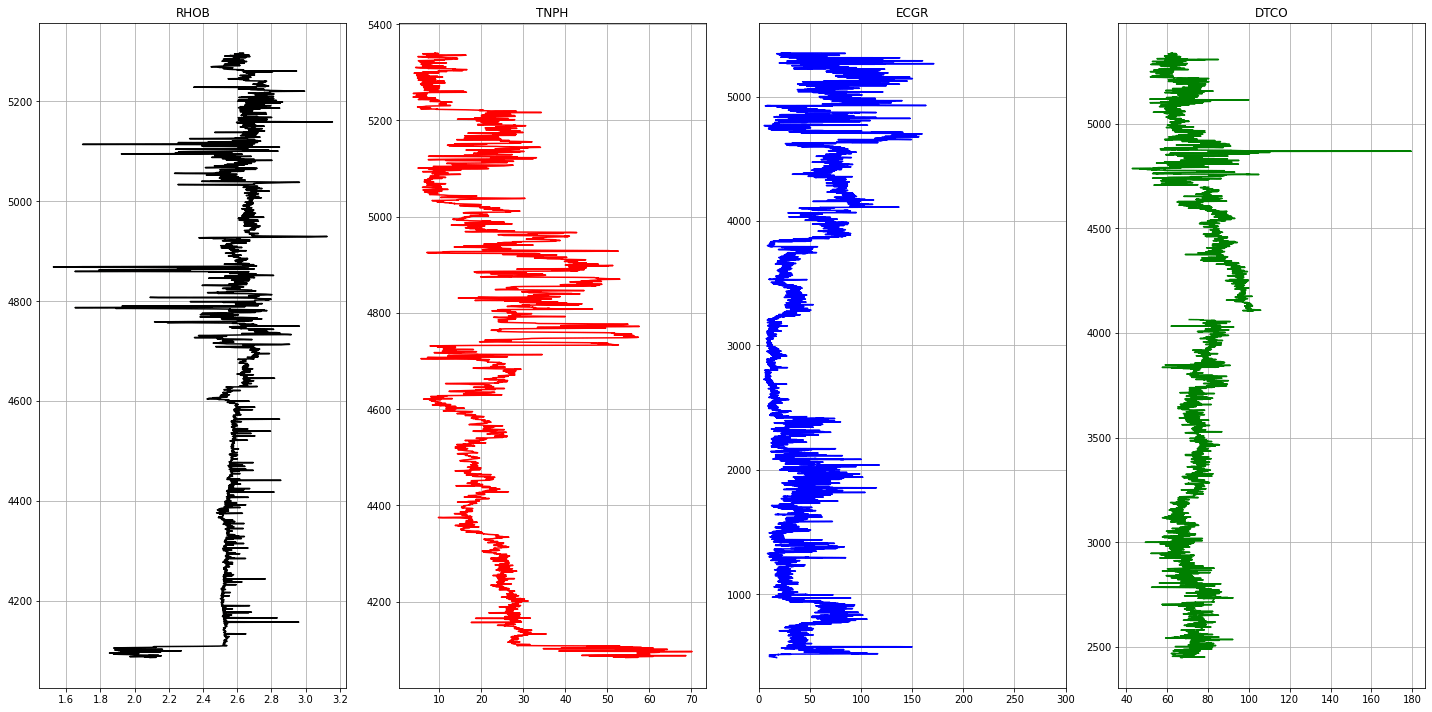
Data Preparation
1# restricting depth ranges per well to reduce the number of missing values.2depth_ranges = {3 'Pharos1' : [4000,5000],4 'PoseidonNorth1' : [3800,4900],5 'Torosa1L' : [3600, 4600],6 'Poseidon2' : [4100, 5000],7 'Proteus1' : [4950, 5200],8 'Kronos1' : [4750,5000],9 'Poseidon1' : [4600, 4950],10 'Boreas1' : [4000,5100]1112}1314for well in well_list :15 df = las_df[well]16 df = df[columns_of_interest].reset_index()17 print("\nWell Name : ", well)18 print("Well Start : ", df['DEPT'].min() , '\t Well Stop :', df['DEPT'].max())19 print("Missing Value % ", 100 * df.isnull().sum()/df.shape[0])2021 depth_start = depth_ranges[well][0]22 depth_stop = depth_ranges[well][-1]2324 condition = (df['DEPT'] >= depth_start) & (df['DEPT'] <=depth_stop)25 las_df[well] = df[condition]2627 assert (las_df[well]['DEPT'].min() >= depth_start) and (las_df[well]['DEPT'].max() <= depth_stop) , 'Depths change error'28 print("New Well Start : ", las_df[well]['DEPT'].min() , '\t New Well Stop :',las_df[well]['DEPT'].max(), '\t Datapoints : ', las_df[well].shape[0])29 print("New Missing Value % ", 100 * las_df[well].isnull().sum()/las_df[well].shape[0])30 print('\n--------------------------------------------------------')
1Well Name : Poseidon12Well Start : 579.5 Well Stop : 5115.53Missing Value % DEPT 0.0000004RHOB 86.2008165TNPH 85.7819916ECGR 17.4804367DTCO 87.5564868dtype: float649New Well Start : 4600.0 New Well Stop : 4950.0 Datapoints : 70110New Missing Value % DEPT 0.011RHOB 0.012TNPH 0.013ECGR 0.014DTCO 0.015dtype: float641617--------------------------------------------------------1819Well Name : Kronos120Well Start : 531.0 Well Stop : 5322.521Missing Value % DEPT 0.00000022RHOB 74.98956623TNPH 75.15651124ECGR 2.49374025DTCO 87.44783026dtype: float6427New Well Start : 4750.0 New Well Stop : 5000.0 Datapoints : 50128New Missing Value % DEPT 0.00000029RHOB 6.18762530TNPH 6.78642731ECGR 0.00000032DTCO 0.00000033dtype: float643435--------------------------------------------------------3637Well Name : Boreas138Well Start : 200.0 Well Stop : 5205.539Missing Value % DEPT 0.00000040RHOB 76.56811841TNPH 76.53815442ECGR 3.01638043DTCO 63.08429944dtype: float6445New Well Start : 4000.0 New Well Stop : 5100.0 Datapoints : 220146New Missing Value % DEPT 0.00000047RHOB 2.08995948TNPH 1.86279049ECGR 4.13448450DTCO 1.13584751dtype: float645253--------------------------------------------------------5455Well Name : Proteus156Well Start : 489.5 Well Stop : 5249.557Missing Value % DEPT 0.00000058RHOB 93.22550259TNPH 92.82638460ECGR 1.89055861DTCO 52.22140562dtype: float6463New Well Start : 4950.0 New Well Stop : 5200.0 Datapoints : 50164New Missing Value % DEPT 0.065RHOB 0.066TNPH 0.067ECGR 0.068DTCO 0.069dtype: float647071--------------------------------------------------------7273Well Name : Pharos174Well Start : 482.0 Well Stop : 5225.575Missing Value % DEPT 0.00000076RHOB 75.23187277TNPH 74.93676278ECGR 0.22133279DTCO 76.11720180dtype: float6481New Well Start : 4000.0 New Well Stop : 5000.0 Datapoints : 200182New Missing Value % DEPT 0.00000083RHOB 0.49975084TNPH 0.54972585ECGR 0.00000086DTCO 5.24737687dtype: float648889--------------------------------------------------------9091Well Name : Torosa1L92Well Start : 555.0 Well Stop : 4683.093Missing Value % DEPT 0.00000094RHOB 73.61027095TNPH 70.81264496ECGR 0.18166497DTCO 7.03645498dtype: float6499New Well Start : 3600.0 New Well Stop : 4600.0 Datapoints : 2001100New Missing Value % DEPT 0.0101RHOB 0.0102TNPH 0.0103ECGR 0.0104DTCO 0.0105dtype: float64106107--------------------------------------------------------108109Well Name : PoseidonNorth1110Well Start : 508.0 Well Stop : 5279.5111Missing Value % DEPT 0.000000112RHOB 75.901090113TNPH 75.869656114ECGR 18.692372115DTCO 67.026404116dtype: float64117New Well Start : 3800.0 New Well Stop : 4900.0 Datapoints : 2201118New Missing Value % DEPT 0.0119RHOB 0.0120TNPH 0.0121ECGR 0.0122DTCO 0.0123dtype: float64124125--------------------------------------------------------126127Well Name : Poseidon2128Well Start : 490.0 Well Stop : 5351.0129Missing Value % DEPT 0.000000130RHOB 75.079708131TNPH 74.154068132ECGR 0.123419133DTCO 41.571531134dtype: float64135New Well Start : 4100.0 New Well Stop : 5000.0 Datapoints : 1801136New Missing Value % DEPT 0.000000137RHOB 0.000000138TNPH 0.000000139ECGR 0.000000140DTCO 1.554692141dtype: float64142143--------------------------------------------------------
1# Concatenating well data into one dataframe2# Adding a column called Well_name to distinguish data points of each well3for well in well_list :4 las_df[well]['Well'] = well56well_df = pd.concat(las_df.values(), axis=0)7well_df = well_df[columns_of_interest + ['Well']]8well_df.head()
| RHOB | TNPH | ECGR | DTCO | Well | |
| 8041 | 2.5211 | 10.10 | 34.3997 | 70.4003 | Poseidon1 |
| 8042 | 2.5112 | 12.40 | 42.8228 | 80.4120 | Poseidon1 |
| 8043 | 2.5277 | 15.30 | 60.1452 | 82.5646 | Poseidon1 |
| 8044 | 2.5127 | 12.28 | 36.4007 | 72.5366 | Poseidon1 |
| 8045 | 2.5271 | 10.16 | 35.3279 | 70.7520 | Poseidon1 |
Missing Value Treatment
1missing_values_df = pd.DataFrame( index = well_df.columns.values )234for well_name in well_df['Well'].unique() :5 condition = well_df['Well'] == well_name6 subset = well_df[condition]7 missing_values_df[well_name] = 100 * subset.isnull().sum()/subset.shape[0]8missing_values_df = missing_values_df.T9missing_values_df.sort_values(by=['DTCO'],ascending=[True]).style.bar()
| RHOB | TNPH | ECGR | DTCO | Well | |
| Poseidon1 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| Kronos1 | 6.187625 | 6.786427 | 0.000000 | 0.000000 | 0.000000 |
| Proteus1 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| Torosa1L | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| PoseidonNorth1 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| Boreas1 | 2.089959 | 1.862790 | 4.134484 | 1.135847 | 0.000000 |
| Poseidon2 | 0.000000 | 0.000000 | 0.000000 | 1.554692 | 0.000000 |
| Pharos1 | 0.499750 | 0.549725 | 0.000000 | 5.247376 | 0.000000 |
Train & Test split
1training_wells = ['PoseidonNorth1', 'Torosa1L','Boreas1', 'Poseidon2','Kronos1', 'Proteus1']2testing_wells = [well for well in well_list if well not in training_wells]34print('Training Wells : ',training_wells)5print('Testing Wells : ' , testing_wells)67train_dataset = pd.concat([las_df[well] for well in training_wells],axis=0)8test_dataset = pd.concat([las_df[well] for well in testing_wells],axis=0)9train_dataset.head()
1Training Wells : ['PoseidonNorth1', 'Torosa1L', 'Boreas1', 'Poseidon2', 'Kronos1', 'Proteus1']2Testing Wells : ['Poseidon1', 'Pharos1']
| DEPT | RHOB | TNPH | ECGR | DTCO | Well | |
| 6584 | 3800.0 | 2.5913 | 35.5945 | 83.2622 | 98.2931 | PoseidonNorth1 |
| 6585 | 3800.5 | 2.5680 | 34.2785 | 73.9849 | 98.1903 | PoseidonNorth1 |
| 6586 | 3801.0 | 2.6482 | 29.6083 | 83.8913 | 98.1472 | PoseidonNorth1 |
| 6587 | 3801.5 | 2.7102 | 34.2985 | 91.5737 | 95.7302 | PoseidonNorth1 |
| 6588 | 3802.0 | 2.9027 | 34.7045 | 86.1709 | 95.8680 | PoseidonNorth1 |
1train_dataset.shape
1(9206, 6)
1100 * train_dataset.isnull().sum() / train_dataset.shape[0]
1DEPT 0.0000002RHOB 0.8364113TNPH 0.8146864ECGR 0.9884865DTCO 0.5757116Well 0.0000007dtype: float64
1train_dataset.dropna(subset=columns_of_interest, inplace=True)2test_dataset.dropna(subset=columns_of_interest, inplace=True)
Correlation
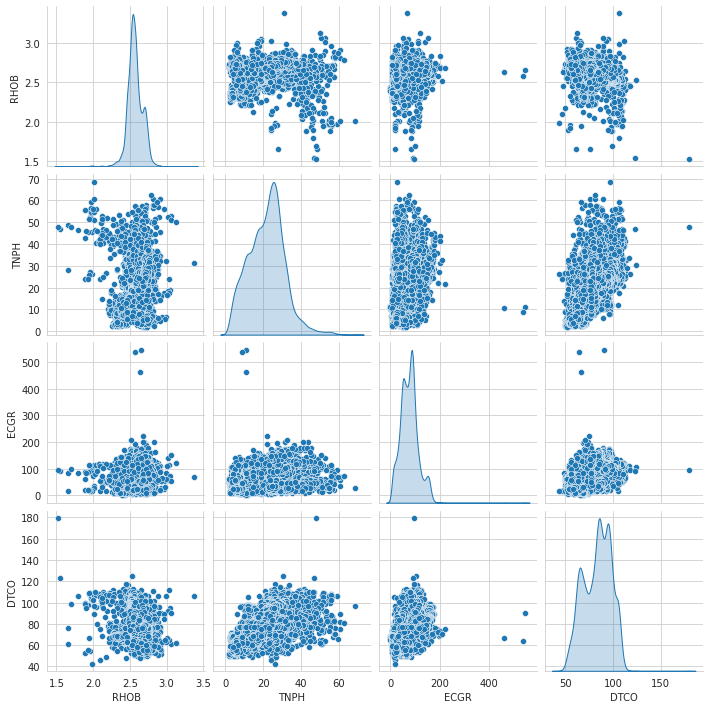
1# Pairplot2import seaborn as sns3sns.set_style('whitegrid')4sns.pairplot(data=train_dataset, vars = ['RHOB', 'TNPH','ECGR','DTCO'], diag_kind='kde')
1<seaborn.axisgrid.PairGrid at 0x7f511c422990>
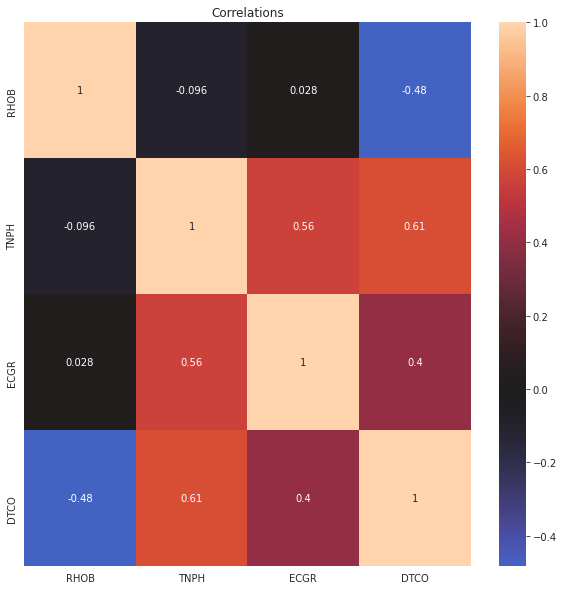
1fig,ax = plt.subplots(figsize=(10,10))2sns.heatmap(train_dataset[columns_of_interest].corr(method='spearman'), annot=True ,ax=ax, center=0)3plt.title('Correlations')4plt.show()
Normalization
1# move the depth column to the right2depth_train, depth_pred = train_dataset.pop('DEPT'), test_dataset.pop('DEPT')3train_dataset['DEPT'], test_dataset['DEPT'] = depth_train, depth_pred
1columns_of_interest
1['RHOB', 'TNPH', 'ECGR', 'DTCO']
1from sklearn.compose import ColumnTransformer2from sklearn.preprocessing import PowerTransformer345scaler = PowerTransformer(method='yeo-johnson')6column_drop = ['WELL','DEPT']7ct = ColumnTransformer([('transform', scaler, columns_of_interest)], remainder='passthrough')89train_dataset_norm = ct.fit_transform(train_dataset)10train_dataset_norm = pd.DataFrame(train_dataset_norm, columns = train_dataset.columns)11train_dataset_norm
| RHOB | TNPH | ECGR | DTCO | Well | DEPT | |
| 0 | 0.194983 | 1.41744 | 0.288585 | 1.069585 | PoseidonNorth1 | 3800.0 |
| 1 | -0.024624 | 1.295228 | 0.0322 | 1.061787 | PoseidonNorth1 | 3800.5 |
| 2 | 0.74861 | 0.851648 | 0.305598 | 1.058519 | PoseidonNorth1 | 3801.0 |
| 3 | 1.380549 | 1.297094 | 0.509886 | 0.876203 | PoseidonNorth1 | 3801.5 |
| 4 | 3.543468 | 1.334914 | 0.366876 | 0.886546 | PoseidonNorth1 | 3802.0 |
| ... | ... | ... | ... | ... | ... | ... |
| 8969 | -3.48235 | -0.712419 | -1.350794 | -1.010424 | Proteus1 | 5198.0 |
| 8970 | 1.775529 | -0.25349 | 0.321077 | -1.018072 | Proteus1 | 5198.5 |
| 8971 | 1.506393 | -0.751591 | -2.436269 | -1.541108 | Proteus1 | 5199.0 |
| 8972 | 2.664714 | -2.337805 | -2.461197 | -1.597139 | Proteus1 | 5199.5 |
| 8973 | 0.283779 | -2.130702 | -1.708333 | -1.674272 | Proteus1 | 5200.0 |
8974 rows × 6 columns
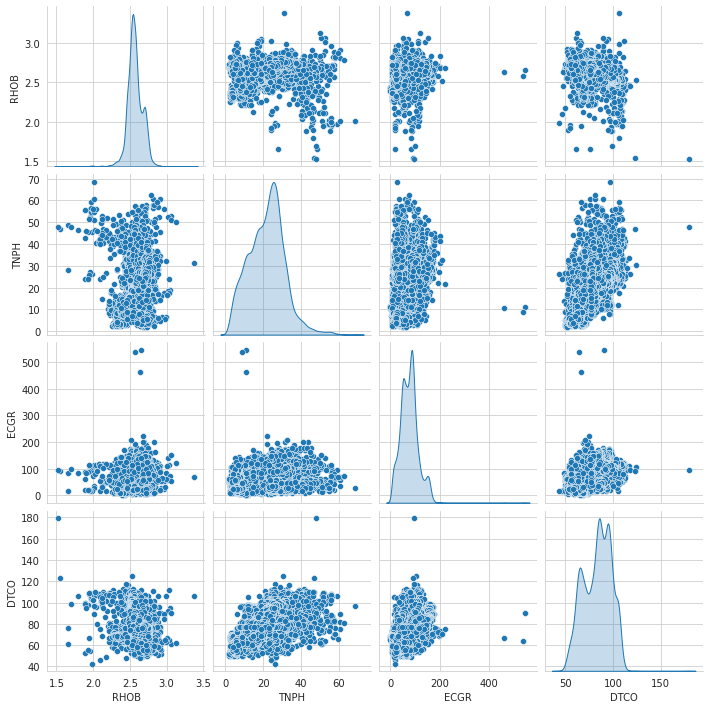
1# Pair Plot after normalization23sns.pairplot(data=train_dataset, vars = ['RHOB', 'TNPH','ECGR','DTCO'], diag_kind='kde')
1<seaborn.axisgrid.PairGrid at 0x7f5102e4e350>
Outlier Removal
1train_dataset_drop = train_dataset_norm.copy()2train_dataset_drop = train_dataset_drop.drop(columns=['Well','DEPT'], )3train_dataset_drop
| RHOB | TNPH | ECGR | DTCO | |
| 0 | 0.194983 | 1.41744 | 0.288585 | 1.069585 |
| 1 | -0.024624 | 1.295228 | 0.0322 | 1.061787 |
| 2 | 0.74861 | 0.851648 | 0.305598 | 1.058519 |
| 3 | 1.380549 | 1.297094 | 0.509886 | 0.876203 |
| 4 | 3.543468 | 1.334914 | 0.366876 | 0.886546 |
| ... | ... | ... | ... | ... |
| 8969 | -3.48235 | -0.712419 | -1.350794 | -1.010424 |
| 8970 | 1.775529 | -0.25349 | 0.321077 | -1.018072 |
| 8971 | 1.506393 | -0.751591 | -2.436269 | -1.541108 |
| 8972 | 2.664714 | -2.337805 | -2.461197 | -1.597139 |
| 8973 | 0.283779 | -2.130702 | -1.708333 | -1.674272 |
8974 rows × 4 columns
1from sklearn.svm import OneClassSVM2svm = OneClassSVM(nu=0.1)3yhat = svm.fit_predict(train_dataset_drop)4mask = yhat != -15train_dataset_svm = train_dataset_norm[mask]6print('Number of points after outliers removed with One-class SVM :', len(train_dataset_svm))
1Number of points after outliers removed with One-class SVM : 8076
Modelling
1y_train = train_dataset_svm['DTCO']2X_train = train_dataset_svm[['RHOB','TNPH','ECGR']]
1# Derived Features2from scipy.ndimage import median_filter34for col in X_train.columns :5 X_train[col + '_GRAD'] = np.gradient(X_train[col].values)6 X_train[col + '_AVG'] = median_filter(X_train[col].values.tolist(),10,mode='nearest')
1X_train.columns
1Index(['RHOB', 'TNPH', 'ECGR', 'RHOB_GRAD', 'RHOB_AVG', 'TNPH_GRAD',2 'TNPH_AVG', 'ECGR_GRAD', 'ECGR_AVG'],3 dtype='object')
1# Random Forest2from sklearn.ensemble import RandomForestRegressor3rf_model = RandomForestRegressor()45rf_model.fit(X_train, y_train)6print('Train Accuracy :', rf_model.score(X_train, y_train))78# for well in test_wells :9# X_test, y_test = process_test_set(test_dataset, well, scaler)10# print('Test Accuracy in ' + well + ' : ', rf_model.score(X_test_scaled, y_test))
1Train Accuracy : 0.9917603654437802
1# Gradient Boost2from sklearn.ensemble import GradientBoostingRegressor3gb_model = GradientBoostingRegressor()4gb_model.fit(X_train, y_train)5print('Train Accuracy :',gb_model.score(X_train, y_train))
1Train Accuracy : 0.9081315988009683
Hyper Parameter Tuning
1# estimator = GradientBoostingRegressor()23# ## Hyperparameters4# max_depth = [10, 20, 30,50, 100]5# min_samples_leaf = [1, 2,3, 4]6# min_samples_split = [2, 10]7# n_estimators = [100,200,300]89# param_grid = {'n_estimators': n_estimators,'max_depth': max_depth , 'min_samples_leaf' : min_samples_leaf, 'min_samples_split' : min_samples_split}1011# from sklearn.model_selection import RandomizedSearchCV12# search = RandomizedSearchCV(estimator, param_grid , cv = 5, verbose=1,n_jobs=-1)13# search.fit(X_train, y_train)14# search.best_params_
1# estimator = RandomForestRegressor()2# ## Hyperparameters3# max_depth = [10, 20, 30,50, 100]4# min_samples_leaf = [1, 2,3, 4]5# min_samples_split = [2, 10]6# n_estimators = [100,200,300]78# param_grid = {'n_estimators': n_estimators,'max_depth': max_depth , 'min_samples_leaf' : min_samples_leaf, 'min_samples_split' : min_samples_split}910# from sklearn.model_selection import RandomizedSearchCV11# search = RandomizedSearchCV(estimator, param_grid , cv = 5, verbose=1,n_jobs=-1, n_iter=20)12# search.fit(X_train, y_train)13# search.best_params_
1#search.best_estimator_
1# New Gradient Boosting model with the best hyper parameters2gb_model_tuned = GradientBoostingRegressor(max_depth = 20, n_estimators= 100,min_samples_leaf=4,min_samples_split=2)3gb_model_tuned.fit(X_train, y_train)4print('Train Accuracy :',gb_model_tuned.score(X_train, y_train))
1Train Accuracy : 0.9998761339778823
1rf_model_tuned = RandomForestRegressor(max_depth=50, min_samples_leaf=2, min_samples_split=10,2 n_estimators=200)3rf_model_tuned.fit(X_train,y_train)
1RandomForestRegressor(max_depth=50, min_samples_leaf=2, min_samples_split=10,2 n_estimators=200)
Test Wells
1# normalisation2test_dataset_norm = pd.DataFrame(ct.transform(test_dataset), columns = test_dataset.columns)
1test_dataset_norm
| RHOB | TNPH | ECGR | DTCO | Well | DEPT | |
| 0 | -0.454429 | -1.257473 | -1.226668 | -0.912366 | Poseidon1 | 4600.0 |
| 1 | -0.543094 | -0.975991 | -0.928349 | -0.233124 | Poseidon1 | 4600.5 |
| 2 | -0.394922 | -0.638131 | -0.372358 | -0.082209 | Poseidon1 | 4601.0 |
| 3 | -0.529706 | -0.990353 | -1.153705 | -0.770638 | Poseidon1 | 4601.5 |
| 4 | -0.400345 | -1.249951 | -1.192646 | -0.889156 | Poseidon1 | 4602.0 |
| ... | ... | ... | ... | ... | ... | ... |
| 2579 | -0.847423 | -1.802829 | -2.111444 | -1.249448 | Pharos1 | 4998.0 |
| 2580 | -0.406668 | -1.645907 | -1.818261 | -1.338187 | Pharos1 | 4998.5 |
| 2581 | -1.341027 | -1.628059 | -1.834552 | -1.27002 | Pharos1 | 4999.0 |
| 2582 | -2.260172 | -2.131092 | -2.367431 | -1.088848 | Pharos1 | 4999.5 |
| 2583 | -2.370962 | -2.157379 | -2.45466 | -0.973624 | Pharos1 | 5000.0 |
2584 rows × 6 columns
1# outlier removal2test_dataset_drop = test_dataset_norm.copy()34yhat = svm.predict(test_dataset_drop[['RHOB','TNPH','ECGR','DTCO']])5mask = yhat != -16test_dataset_svm = test_dataset_norm[mask]7test_dataset_svm_unscaled = test_dataset[mask]8print('Number of points after outliers removed with One-class SVM :', len(test_dataset_svm))
1Number of points after outliers removed with One-class SVM : 2152
1X_test = []2y_test = []34for i, well in enumerate(testing_wells) :5 test = None6 test = test_dataset_svm.loc[test_dataset_svm['Well'] == well]78 test = test.drop(columns=['Well','DEPT'])91011 # Derived Features1213 for col in ['RHOB','TNPH','ECGR'] :14 test[col + '_GRAD'] = np.gradient(test[col].values)15 test[col + '_AVG'] = median_filter(test[col].values.tolist(),10,mode='nearest')1617 y_test.append(test.pop('DTCO').values)18 X_test.append(test.values)
1# predictions2from sklearn.metrics import mean_squared_error3y_test_pred_gb = []4y_test_pred_rf = []5for i in range(len(X_test)) :6 y_test_pred_gb.append(gb_model_tuned.predict(X_test[i]))7 y_test_pred_rf.append(rf_model_tuned.predict(X_test[i]))8 print('\n------------------------')9 print('Test Accuracy of Well using Gradient Boosting Model ', testing_wells[i], ' : ', gb_model_tuned.score(X_test[i], y_test[i]))10 print('MSE of Well using Gradient Boosting Model ', testing_wells[i], ' : ', mean_squared_error(y_test[i], y_test_pred_gb[i]))11 print('Test Accuracy of Well using RF Model ', testing_wells[i], ' : ', rf_model_tuned.score(X_test[i], y_test[i]))12 print('MSE of Well using RF Model ', testing_wells[i], ' : ', mean_squared_error(y_test[i], y_test_pred_rf[i]))
1------------------------2Test Accuracy of Well using Gradient Boosting Model Poseidon1 : 0.75192031138516713MSE of Well using Gradient Boosting Model Poseidon1 : 0.1628512611839054Test Accuracy of Well using RF Model Poseidon1 : 0.78129186139131315MSE of Well using RF Model Poseidon1 : 0.1435703841877500367------------------------8Test Accuracy of Well using Gradient Boosting Model Pharos1 : 0.75457019418061169MSE of Well using Gradient Boosting Model Pharos1 : 0.0978798512903486510Test Accuracy of Well using RF Model Pharos1 : 0.786618796747500811MSE of Well using RF Model Pharos1 : 0.08509854935011467
Using Artificial Neural Networks
1import tensorflow as tf2tf.random.set_seed(42)3import keras4from keras.models import Sequential5from keras.layers import Dense, Dropout, Activation, BatchNormalization, Dropout6from keras.callbacks import ModelCheckpoint
12021-07-11 19:25:27.814036: W tensorflow/stream_executor/platform/default/dso_loader.cc:60] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory22021-07-11 19:25:27.814077: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
1# Over Parameterised ANN2model = Sequential([3 Dense(128, activation='relu',kernel_initializer='normal', input_shape=(9,) ) ,4 BatchNormalization(),5 Dropout(0.10),67 Dense(64, kernel_initializer='normal',activation='relu' ) ,8 BatchNormalization(),9 Dropout(0.10),1011 Dense(32, kernel_initializer='normal',activation='relu' ) ,12 BatchNormalization(),13 Dropout(0.10),1415 Dense(16, kernel_initializer='normal',activation='relu' ) ,16 BatchNormalization(),17 Dropout(0.10),1819 Dense(8, kernel_initializer='normal',activation='relu' ) ,20 BatchNormalization(),21 Dropout(0.10),2223 Dense(4, kernel_initializer='normal',activation='relu' ) ,24 BatchNormalization(),25 Dropout(0.10),2627 Dense(units=1, kernel_initializer='normal') ]2829)30313233model.compile(optimizer='adam', loss='mean_squared_error')34print(model.summary())35model_checkpoint = ModelCheckpoint('best_model',monitor='val_loss', mode = 'min', save_best_only=True, verbose=1)3637def scheduler(epoch, lr):38 if epoch <=10 :39 return lr40 else :41 return lr * tf.math.exp(-0.1 * epoch)4243lr_callback = tf.keras.callbacks.LearningRateScheduler(scheduler)4445X_train_tf = tf.convert_to_tensor(X_train.values, np.float32)46y_train_tf = tf.convert_to_tensor(y_train.values, np.float32)
12021-07-11 19:25:32.081060: I tensorflow/compiler/jit/xla_cpu_device.cc:41] Not creating XLA devices, tf_xla_enable_xla_devices not set22021-07-11 19:25:32.081302: W tensorflow/stream_executor/platform/default/dso_loader.cc:60] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory32021-07-11 19:25:32.081316: W tensorflow/stream_executor/cuda/cuda_driver.cc:326] failed call to cuInit: UNKNOWN ERROR (303)42021-07-11 19:25:32.081333: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (p-fbd0b10b-3636-4896-bb87-fbf506835cad): /proc/driver/nvidia/version does not exist52021-07-11 19:25:32.081543: I tensorflow/core/platform/cpu_feature_guard.cc:142] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA6To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.72021-07-11 19:25:32.081688: I tensorflow/compiler/jit/xla_gpu_device.cc:99] Not creating XLA devices, tf_xla_enable_xla_devices not set8Model: "sequential"9_________________________________________________________________10Layer (type) Output Shape Param #11=================================================================12dense (Dense) (None, 128) 128013_________________________________________________________________14batch_normalization (BatchNo (None, 128) 51215_________________________________________________________________16dropout (Dropout) (None, 128) 017_________________________________________________________________18dense_1 (Dense) (None, 64) 825619_________________________________________________________________20batch_normalization_1 (Batch (None, 64) 25621_________________________________________________________________22dropout_1 (Dropout) (None, 64) 023_________________________________________________________________24dense_2 (Dense) (None, 32) 208025_________________________________________________________________26batch_normalization_2 (Batch (None, 32) 12827_________________________________________________________________28dropout_2 (Dropout) (None, 32) 029_________________________________________________________________30dense_3 (Dense) (None, 16) 52831_________________________________________________________________32batch_normalization_3 (Batch (None, 16) 6433_________________________________________________________________34dropout_3 (Dropout) (None, 16) 035_________________________________________________________________36dense_4 (Dense) (None, 8) 13637_________________________________________________________________38batch_normalization_4 (Batch (None, 8) 3239_________________________________________________________________40dropout_4 (Dropout) (None, 8) 041_________________________________________________________________42dense_5 (Dense) (None, 4) 3643_________________________________________________________________44batch_normalization_5 (Batch (None, 4) 1645_________________________________________________________________46dropout_5 (Dropout) (None, 4) 047_________________________________________________________________48dense_6 (Dense) (None, 1) 549=================================================================50Total params: 13,32951Trainable params: 12,82552Non-trainable params: 50453_________________________________________________________________54None
1history = model.fit(X_train_tf, y_train_tf, batch_size=00, validation_split=0.3, epochs=40,callbacks=[model_checkpoint, lr_callback])
1Epoch 1/4022021-07-11 19:25:32.416134: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:116] None of the MLIR optimization passes are enabled (registered 2)32021-07-11 19:25:32.416620: I tensorflow/core/platform/profile_utils/cpu_utils.cc:112] CPU Frequency: 2499995000 Hz4177/177 [==============================] - 2s 6ms/step - loss: 0.6706 - val_loss: 1.010956Epoch 00001: val_loss improved from inf to 1.01087, saving model to best_model72021-07-11 19:25:35.935800: W tensorflow/python/util/util.cc:348] Sets are not currently considered sequences, but this may change in the future, so consider avoiding using them.8INFO:tensorflow:Assets written to: best_model/assets9Epoch 2/4010177/177 [==============================] - 1s 3ms/step - loss: 0.2181 - val_loss: 0.67171112Epoch 00002: val_loss improved from 1.01087 to 0.67171, saving model to best_model13INFO:tensorflow:Assets written to: best_model/assets14Epoch 3/4015177/177 [==============================] - 1s 3ms/step - loss: 0.1860 - val_loss: 0.21831617Epoch 00003: val_loss improved from 0.67171 to 0.21830, saving model to best_model18INFO:tensorflow:Assets written to: best_model/assets19Epoch 4/4020177/177 [==============================] - 1s 3ms/step - loss: 0.1927 - val_loss: 0.12772122Epoch 00004: val_loss improved from 0.21830 to 0.12774, saving model to best_model23INFO:tensorflow:Assets written to: best_model/assets24Epoch 5/4025177/177 [==============================] - 1s 4ms/step - loss: 0.1761 - val_loss: 0.13952627Epoch 00005: val_loss did not improve from 0.1277428Epoch 6/4029177/177 [==============================] - 1s 3ms/step - loss: 0.1792 - val_loss: 0.14093031Epoch 00006: val_loss did not improve from 0.1277432Epoch 7/4033177/177 [==============================] - 1s 3ms/step - loss: 0.1682 - val_loss: 0.17903435Epoch 00007: val_loss did not improve from 0.1277436Epoch 8/4037177/177 [==============================] - 1s 3ms/step - loss: 0.1706 - val_loss: 0.14033839Epoch 00008: val_loss did not improve from 0.1277440Epoch 9/4041177/177 [==============================] - 1s 3ms/step - loss: 0.1751 - val_loss: 0.13924243Epoch 00009: val_loss did not improve from 0.1277444Epoch 10/4045177/177 [==============================] - 1s 3ms/step - loss: 0.1725 - val_loss: 0.15804647Epoch 00010: val_loss did not improve from 0.1277448Epoch 11/4049177/177 [==============================] - 1s 3ms/step - loss: 0.1775 - val_loss: 0.13405051Epoch 00011: val_loss did not improve from 0.1277452Epoch 12/4053177/177 [==============================] - 1s 3ms/step - loss: 0.1675 - val_loss: 0.13675455Epoch 00012: val_loss did not improve from 0.1277456Epoch 13/4057177/177 [==============================] - 1s 3ms/step - loss: 0.1543 - val_loss: 0.14065859Epoch 00013: val_loss did not improve from 0.1277460Epoch 14/4061177/177 [==============================] - 1s 3ms/step - loss: 0.1471 - val_loss: 0.14366263Epoch 00014: val_loss did not improve from 0.1277464Epoch 15/4065177/177 [==============================] - 1s 4ms/step - loss: 0.1496 - val_loss: 0.14326667Epoch 00015: val_loss did not improve from 0.1277468Epoch 16/4069177/177 [==============================] - 1s 3ms/step - loss: 0.1599 - val_loss: 0.14237071Epoch 00016: val_loss did not improve from 0.1277472Epoch 17/4073177/177 [==============================] - 1s 3ms/step - loss: 0.1613 - val_loss: 0.14137475Epoch 00017: val_loss did not improve from 0.1277476Epoch 18/4077177/177 [==============================] - 1s 3ms/step - loss: 0.1445 - val_loss: 0.14217879Epoch 00018: val_loss did not improve from 0.1277480Epoch 19/4081177/177 [==============================] - 1s 3ms/step - loss: 0.1512 - val_loss: 0.14208283Epoch 00019: val_loss did not improve from 0.1277484Epoch 20/4085177/177 [==============================] - 1s 3ms/step - loss: 0.1469 - val_loss: 0.14168687Epoch 00020: val_loss did not improve from 0.1277488Epoch 21/4089177/177 [==============================] - 1s 3ms/step - loss: 0.1556 - val_loss: 0.14229091Epoch 00021: val_loss did not improve from 0.1277492Epoch 22/4093177/177 [==============================] - 1s 3ms/step - loss: 0.1545 - val_loss: 0.14259495Epoch 00022: val_loss did not improve from 0.1277496Epoch 23/4097177/177 [==============================] - 1s 3ms/step - loss: 0.1496 - val_loss: 0.14269899Epoch 00023: val_loss did not improve from 0.12774100Epoch 24/40101177/177 [==============================] - 1s 3ms/step - loss: 0.1540 - val_loss: 0.1418102103Epoch 00024: val_loss did not improve from 0.12774104Epoch 25/40105177/177 [==============================] - 1s 3ms/step - loss: 0.1472 - val_loss: 0.1407106107Epoch 00025: val_loss did not improve from 0.12774108Epoch 26/40109177/177 [==============================] - 1s 3ms/step - loss: 0.1550 - val_loss: 0.1411110111Epoch 00026: val_loss did not improve from 0.12774112Epoch 27/40113177/177 [==============================] - 1s 3ms/step - loss: 0.1572 - val_loss: 0.1428114115Epoch 00027: val_loss did not improve from 0.12774116Epoch 28/40117177/177 [==============================] - 1s 3ms/step - loss: 0.1663 - val_loss: 0.1426118119Epoch 00028: val_loss did not improve from 0.12774120Epoch 29/40121177/177 [==============================] - 1s 3ms/step - loss: 0.1568 - val_loss: 0.1418122123Epoch 00029: val_loss did not improve from 0.12774124Epoch 30/40125177/177 [==============================] - 1s 3ms/step - loss: 0.1565 - val_loss: 0.1426126127Epoch 00030: val_loss did not improve from 0.12774128Epoch 31/40129177/177 [==============================] - 1s 3ms/step - loss: 0.1573 - val_loss: 0.1442130131Epoch 00031: val_loss did not improve from 0.12774132Epoch 32/40133177/177 [==============================] - 1s 3ms/step - loss: 0.1512 - val_loss: 0.1425134135Epoch 00032: val_loss did not improve from 0.12774136Epoch 33/40137177/177 [==============================] - 1s 4ms/step - loss: 0.1498 - val_loss: 0.1419138139Epoch 00033: val_loss did not improve from 0.12774140Epoch 34/40141177/177 [==============================] - 1s 3ms/step - loss: 0.1529 - val_loss: 0.1418142143Epoch 00034: val_loss did not improve from 0.12774144Epoch 35/40145177/177 [==============================] - 1s 3ms/step - loss: 0.1542 - val_loss: 0.1419146147Epoch 00035: val_loss did not improve from 0.12774148Epoch 36/40149177/177 [==============================] - 1s 3ms/step - loss: 0.1591 - val_loss: 0.1432150151Epoch 00036: val_loss did not improve from 0.12774152Epoch 37/40153177/177 [==============================] - 1s 3ms/step - loss: 0.1611 - val_loss: 0.1430154155Epoch 00037: val_loss did not improve from 0.12774156Epoch 38/40157177/177 [==============================] - 1s 3ms/step - loss: 0.1492 - val_loss: 0.1423158159Epoch 00038: val_loss did not improve from 0.12774160Epoch 39/40161177/177 [==============================] - 1s 3ms/step - loss: 0.1604 - val_loss: 0.1420162163Epoch 00039: val_loss did not improve from 0.12774164Epoch 40/40165177/177 [==============================] - 1s 3ms/step - loss: 0.1460 - val_loss: 0.1428166167Epoch 00040: val_loss did not improve from 0.12774
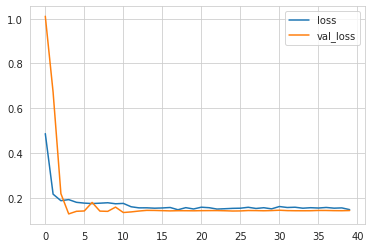
1def plot_model_history(history):2 fig, axes = plt.subplots(nrows=1, ncols=1)3 axes.plot(history.history['loss'])4 axes.plot(history.history['val_loss'])5 axes.legend(['loss','val_loss'])67plot_model_history(history)
1y_test_pred_nn = []23for i in range(len(X_test)) :4 print('Test Well : ',testing_wells[i] )5 X_test_tf = tf.convert_to_tensor(X_test[i], np.float32)6 y_test_tf =tf.convert_to_tensor(y_test[i], np.float32)7 y_test_pred_nn.append(model.predict(X_test_tf))8 print('MSE :', model.evaluate(X_test_tf, y_test_tf))9 print('\n---------------------\n')
1Test Well : Poseidon1216/16 [==============================] - 0s 1ms/step - loss: 0.16553MSE : 0.1655042469501495445---------------------67Test Well : Pharos1852/52 [==============================] - 0s 883us/step - loss: 0.07439MSE : 0.074273362755775451011---------------------
1#2for i in range(len(testing_wells)) :3 preds = y_test_pred_nn[i]4 rows = preds.shape[0]56 df = pd.DataFrame(columns = columns_of_interest, index = range(rows))7 for j in range(len(columns_of_interest)-1) :8 df[:][j] = 09 df['DTCO'] = preds1011 rescaled_df = pd.DataFrame(ct.named_transformers_.transform.inverse_transform(df) , columns = columns_of_interest)121314 condition = test_dataset_svm_unscaled['Well'] == testing_wells[i]1516 test_dataset_svm_unscaled.loc[condition, 'DTCO_Predicted'] = rescaled_df['DTCO'].values
1test_dataset_svm_unscaled
| RHOB | TNPH | ECGR | DTCO | Well | DEPT | DTCO_Predicted | |
| 8041 | 2.5211 | 10.1000 | 34.3997 | 70.4003 | Poseidon1 | 4600.0 | 73.254893 |
| 8042 | 2.5112 | 12.4000 | 42.8228 | 80.4120 | Poseidon1 | 4600.5 | 72.147674 |
| 8043 | 2.5277 | 15.3000 | 60.1452 | 82.5646 | Poseidon1 | 4601.0 | 75.254447 |
| 8044 | 2.5127 | 12.2800 | 36.4007 | 72.5366 | Poseidon1 | 4601.5 | 72.460500 |
| 8045 | 2.5271 | 10.1600 | 35.3279 | 70.7520 | Poseidon1 | 4602.0 | 76.343241 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 9030 | 2.4565 | 6.8472 | 16.4082 | 64.9338 | Pharos1 | 4997.0 | 65.889312 |
| 9031 | 2.4493 | 6.0554 | 13.1265 | 64.0184 | Pharos1 | 4997.5 | 65.930052 |
| 9032 | 2.4766 | 5.9867 | 13.4912 | 65.2055 | Pharos1 | 4998.0 | 66.617091 |
| 9033 | 2.5264 | 7.1198 | 19.6898 | 63.8091 | Pharos1 | 4998.5 | 66.574747 |
| 9034 | 2.4183 | 7.2514 | 19.3252 | 64.8829 | Pharos1 | 4999.0 | 66.289676 |
2152 rows × 7 columns
Plotting Results
1# Plotting Results2def plot_logs(well) :3 columns = columns_of_interest + ['DTCO_Predicted']4 fig,ax = plt.subplots(nrows=1, ncols= len(columns), figsize=(20,10))5 colors = ['black', 'red', 'blue', 'green', 'purple', 'black', 'orange']67 for i,log in enumerate(columns) :8 if log not in ['DEPT','Well'] :9 ax[i].plot(well[log], well['DEPT'], color = colors[i])10 ax[i].set_title(log)11 ax[i].grid(True)1213 ax[2].set_xlim(0,300)14 plt.tight_layout(1.1)15 plt.show()1617for well in testing_wells :18 print('Well Name :', well)19 condition = test_dataset_svm_unscaled['Well'] == well2021 plot_logs(test_dataset_svm_unscaled[condition])
1Well Name : Poseidon1
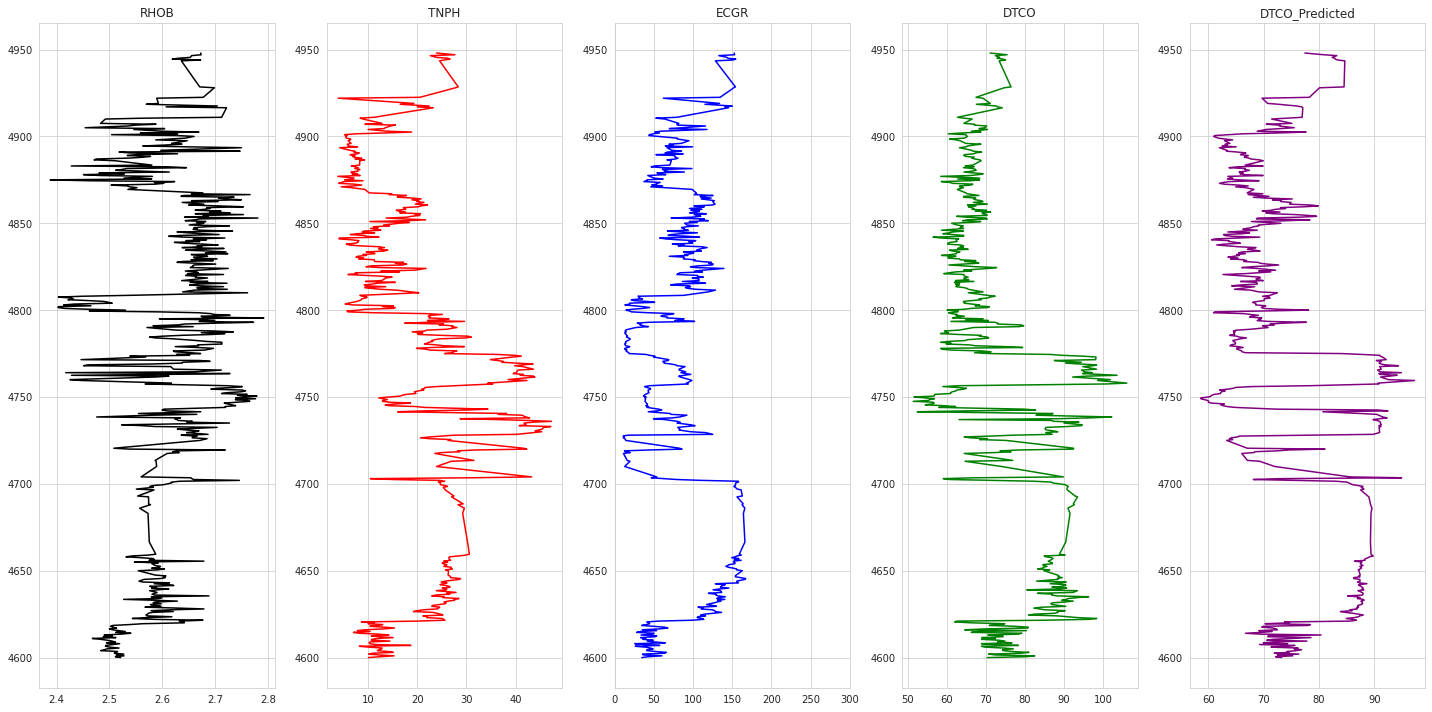
1Well Name : Pharos1